|
To stay alive, bacteria, like other living cells, transduce energy. Two of its main sources are the ATP molecule, and the electrochemical gradients of ions, often called the ion motive forces (IMF). In most bacteria, the dominant IMF is the proton motive force, while in some it is the sodium motive force. To understand IMFs, one needs to consider their electrical nature, which, because bacteria are unicellular, is tightly coupled with all the other processes in the cell. For example, the electrical potential across the membrane is generated by the charge accumulated at the membrane, which drives all ions. However, ions are also driven by specific chemical concentration differences, where the exact concentration of ions in the cell matters as well, particularly of protons. Lastly, bacteria maintain significant osmotic (turgor) pressures, which depend on the difference between the extracellular and intracellular concentrations of all solutes, including ions. The resulting intertwined nature of these physiological variables challenges intuition but is fundamentally important. Bacterial flagellar motor can be used as a single-cell voltmeter. The bacterial cell can be represented as an electric circuit, where the respiration complex plays the role of the imperfect battery with an internal resistance, Ri. The battery is the theoretical maximum voltage that can be obtained from a given carbon source once it is internalised and Ri is the loss from that maximum due to metabolism. The membrane, with different ion proteins (antiporters, symporters, channels, etc.) functions as an external resistance and a capacitance connected in parallel. We are approaching the problem by developing both experimental methods and theoretical frameworks. For example, the experimental assays for measuring ATP concentration, ATP:ADP ratio, ionic strength, membrane potential, pH, intracellular potassium and sodium concentrations, PMF, and SMF. The assays are suitable for both population and single-cell level measurements, and measurements of more than one variable concurrently. Because the effects of ion fluxes are nontrivially intertwined, we are developing pump-leak equations-based models to understand bacterial electrophysiology and free energy fluxes better. 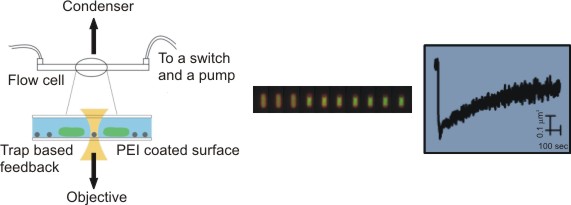 Single bacterial cells are observed during osmotic chalanges. A sequence of images obtained is converted to a high-resolution temporal trace showing changes in cellular volume. Watch the movie of the sequence of fluorescent images shown in the middle. While focused mostly on fundamental science, we have a significant track record of effectively translating our research group findings for industry application. For example, we have worked with Fujifilm Diosynth Biotechnologies, INEOS Bio, Novonesis, GreenBiologics, and WEST Brewery on exploiting bacterial responses to osmotic changes to solve their industrial challenges. In February 2020 Alex McVey and Teuta co-founded a start-up company, OGI Bio Ltd, whose vision is to revolutionize current practices in microbial culturing through modular automation. Four former lab members have worked or continue to work for the company. James Flewellen, with several others in the lab, is currently working on a spinout company whose vision is to fundamentally change the way we sense our environment.   Left: Our home-built microscope and optical trapping system Right: Our prototype microbioreactor, for commercial version see OGI Bio Ltd and speak to Alex McVey. References:(1) Lo WC, Krasnopeeva E, Pilizota T** 'Bacterial electrophysiology' Annual Review of biophysics 2024(2) Krasnopeeva E, Barboza-Perez U E, Rosko J, Pilizota T**, Lo C J** 'Bacterial Flagellar Motor as a Multimodal Biosensor' Methods 2021; 193:5-15 (3) Krasnopeeva E, Lo CJ, Pilizota T**. Single-cell bacterial electrophysiology reveals mechanisms of stress induced damage. Biophys J 2019;116(12): 2390-2399 (4) Buda R*, Liu Y*, Yang J*, Hegde S*, Stevenson K, Bai F** and Pilizota T**. Dynamics of Escherichia coli's passive response to a sudden decrease in external osmolarity. PNAS September 2016, doi:10.1073/pnas.1522185113 (5) Pilizota T, Shaevitz JW. Plasmolysis and cell shape depend on solute outer-membrane permeability during hyperosmotic shock in Escherichia coli Biophys J 18 June 2013, 104(12):2733-2742 Videos:

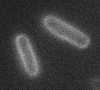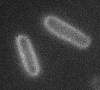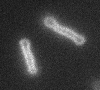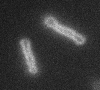
Left: A single cell exposed to an increase in external concentration caused by the addition of NaCl to the media. Red marks the total cell volume and green is the cytoplasmic cell volume. Middle: Cells are exposed to an increase in external osmolarity and observed during recovery. Cytoplasmic cell volume is shown. Faster than real time. Right: Cells are exposed to a very large increase in external sucrose concentration. Total cell volume is shown. 
Left: The bead assay used to detect rotation of the bacterial flagellar motor. In this case, a bead pair is attached to the shortened flagellar filament and thus rotated by the motor Right: This video is courtesy of Keiichi Namba. It illustrates the principle behind the bead assay and back focal plane interferometry, both used to detect the rotation of the bacterial flagellar motor. |
|

 |
DHTML and javascript menus courtesy of
TwinHelix Designs
maintained by Teuta Pilizota last updated: 19th October 2024 |